Building a COVID-19 Research Engine
The COVID-19 Open Research Dataset Challenge dataset was created on Kaggle by a coalition of research groups to encourage Kagglers to help find solutions to the COVID-19 pandemic by making it easier to search thousands of medical research papers. This notebook was my contribution to the challenge
This notebook contains a library called the CORD Research Engine, which uses search and NLP technology to simplify searching the CORD Research Paper dataset for information that would help solve the current pandemic"
Getting Started
Data files
The CORD-19 dataset on Kaggle contains over 59,000 research papers. This notebook references about 2,000 of those papers, located in the directory data. Inside the data directory, the CORD Research data is located in the CORD-19-research-challenge subdirectory. For the full dataset please see the competition page on Kaggle.
Navigating the data
The original notebook was a Python kernel on Kaggle. On kaggle the data directory is /kaggle/input and when you launch a new notebook you are given a block of code to view the data in the directory.
import os
for dirname, _, filenames in os.walk('/kaggle/input'):
for filename in filenames:
print(os.path.join(dirname, filename))The Data Directory
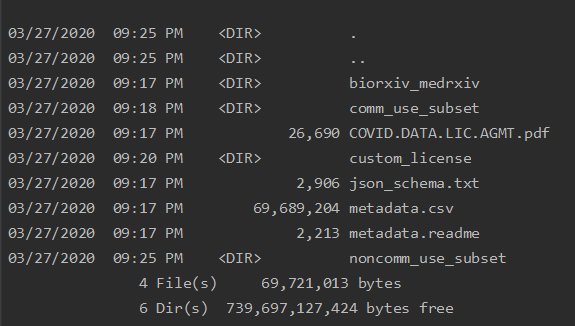
Instead of the code above we will use pathlib for navigating the filesystem because it is a little bit easier to use than os.path. From pathlib we will use the Path and PurePath classes. The Path class provides almost all the functionality we need except that it cannot be passed directly into _pandas.readcsv as can be done with the PurePath.
Now we can create a Path object at the location of the CORD research data and view the contents of that directory.
from pathlib import Path, PurePath
data_path = Path('data/CORD-19-research-challenge')
list(data_path.glob('*'))
The CORD-19 data directory contains four sub-directories, the JSON contents of the research papers, and a metadata.csv file with the metadata for all the research papers. For the actual details of the CORD Research papers dataset, see the description on the Kaggle site.
Research Engine Design
A tale of two indexes
We will build two indexes into the data: one for search, and one for similarity. An index is just a data structure that provides a way to sort some data. In our case, for search, we want to provide a search term to the index and have it return a list of the research papers, sorted by most relevant to the search term.
With the similarity index, the goal is to provide a research paper as input and have the index return a list of papers sorted by the most similar to that paper.
In the real world, and for applications that you may write, you may encounter those two use cases, so we will show the best techniques to fulfill the requirements. Search and Similarity are related but not identical areas, and both the indexes we will build are better at one than the other.
Object-Oriented Design
An object-oriented design is rarely used with notebooks, and for good reason. A notebook is a hybrid of a Python script and a layout template, which both operate in a top to bottom way. Objects are usually meant to be defined in a certain location or context and interacted with multiple times during the course of a program. This is exactly what we want to do here; we intend to create a Research Engine and use it to find research papers during the course of a single session.
There are two main objects in our design: ResearchPapers and Paper. The ResearchPapers class will load and maintain the list of research papers, while Paper will allow us to work with a single research paper.
class ResearchPapers:
def __init__(self, metadata, data_dir='data'):
pass
class Paper:
def __init__(self):
pass
Loading Research Papers
We start with the metadata.csv since this contains the master list of all the research papers and important information about each paper. Some Kaggle kernels started by loading the research papers from the JSON files, but the metadata is considerably smaller, and starting with the metadata might allow us to only load a research paper from a JSON file if required, thereby saving significantly on computational resources.
Load Metadata
The load_metadata function is straightforward - it uses pandas.read_csv() to load the metadata.csv file. Along the way it changes the data types for the Microsoft Academic Paper ID and _pubmedid columns. It also renames a couple of columns, which is optional, and only to make browsing the data a little easier.
@staticmethod
def load_metadata(data_path=None):
if not data_path:
data_path = find_data_dir()
print('Loading metadata from', data_path)
metadata_path = PurePath(data_path) / 'metadata.csv'
dtypes = {'Microsoft Academic Paper ID': 'str', 'pubmed_id': str}
renames = {'source_x': 'source', 'has_full_text': 'has_text'}
# Load the metadata using read.csv
metadata = pd.read_csv(metadata_path, dtype=dtypes,
low_memory=False,
parse_dates=['publish_time']).rename(columns=renames)
metadata = clean_metadata(metadata)
return metadata
Within load_metadata we call clean_metadata. This function is a data cleaning pipeline that fixes some of the issues with the Kaggle data by using a pipeline of data cleaning functions. Notice that each function is specfic to a task, and named specifically for that task, which makes it simple to remove or add cleaning functions. This is important since a new version of the CORD research data is released every week, with new data fixes, so if you can quickly modify your data pipeline, it gives you an advantage.
Clean Metadata
def clean_metadata(metadata):
print('Cleaning metadata')
return metadata.pipe(start) \
.pipe(clean_title) \
.pipe(clean_abstract) \
.pipe(rename_publish_time) \
.pipe(add_date_diff) \
.pipe(drop_missing) \
.pipe(fill_nulls) \
.pipe(apply_tags)
Each function in the clean_metadata pipeline accepts a dataframe and returns the dataframe after modification. The functions are connected by the pandas dataframe pipe function, which is designed for this use case of connecting functions sequentially. At the start of the pipeline is a special function called start which is defined as:
def start(data):
return data.copy()
This returns a copy of the data at the start of a data pipeline. This is very important; it makes sure that the initial data is unchanged and enables you to rerun your notebook from any point without worrying that your data has changed in a way that breaks it. This is a pattern I learned from the presentation Untitled12.ipynb by Vincent D Warmerdam at PyData Eindhoven 2019
Each subsequent function in the pipeline accepts a dataframe and returns a dataframe. Here is clean_title:
def clean_title(data):
# Set junk titles to ''
title_relevant = data.title.fillna('').str.match(_relevant_re_, case=False)
title_short = data.title.fillna('').apply(len) < 30
title_junk = title_short & ~title_relevant
data.loc[title_junk, 'title'] = ''
return data
Load function
After loading the metadata, we have the load function. This is the static function on the ResearchPapers class that actually creates the ResearchPapers instance using ResearchPapers.load().
@classmethod
def load(cls, index=None):
data_path = find_data_dir()
metadata = cls.load_metadata(data_path)
return cls(metadata, data_path, index=index)
Preprocessing
While the clean_metadata function does some text preprocessing, it does that in relation to the metadata.csv file that is released by Kaggle to remove any data issues it might have. We still need to do text preprocessing to prepare the text of the research papers for storing in the search and similarity indexes. Text preprocessing is a necessary part of NLP projects, and, despite the fact that there are a lot of utilities available in NLP libraries to preprocess test, it is still part of the art of being an NLP practictioner. For example, the regex below was hand-crafted with input from Google searches, and a lot of trial and error. I found, for example, that the text prepeocessing that came with gensim was hand-rolled. You may or may not need to follow a similar process for your own text preprocessing, depending on your judgement.
TOKEN_PATTERN = re.compile('^(20|19)\d{2}|(?=[A-Z])[\w\-\d]+$', re.IGNORECASE)
def replace_punctuation(text):
t = re.sub('\(|\)|:|,|;|\.|’|”|“|\?|%|>|<|≥|≤|~|`', '', text)
t = re.sub('/', ' ', t)
t = t.replace("'", '')
return t
def clean(text):
t = text.lower()
t = replace_punctuation(t)
return t
def tokenize(text):
words = nltk.word_tokenize(text)
return [word for word in words
if len(word) > 1
and not word in SIMPLE_STOPWORDS
and TOKEN_PATTERN.match(word)
]
def preprocess(text):
t = clean(text)
tokens = tokenize(t)
return tokens
The preprocess function
Regardless, every NLP project requires a preprocess or equivalent function. Our preprocess function converts the text to lowercase, removes punctuation, and converts the text to tokens. What is very important is that the identical preprocess function be used when preparing the text in batch mode as when using it in query mode. So the preprocess function will be used again on each search query to match accurately against what is stored in the index.
Utility Functions
Parallel Processing
As of the May 2nd data release the CORD Research Paper dataset was over 8GB in size, with more than 59,000 metadata records and well over 60,000 research papers on disk. In order to process the data in a reasonable amount of time, we added a utility function that will run a given function on a list of data using as many cores that are available.
def parallel(func, arr: Collection, max_workers: int = None, leave=False):
"Call `func` on every element of `arr` in parallel using `max_workers`."
max_workers = ifnone(max_workers, multiprocessing.cpu_count())
progress_bar = tqdm(arr)
with ThreadPoolExecutor(max_workers=max_workers) as ex:
futures_to_index = {ex.submit(func, o): i for i, o in enumerate(arr)}
results = []
for f in as_completed(futures_to_index):
results.append((futures_to_index[f], f.result()))
progress_bar.update()
for n in range(progress_bar.n, progress_bar.total):
time.sleep(0.1)
progress_bar.update()
results.sort(key=lambda x: x[0])
return [result for i, result in results]
For example, if we have a list of JSON files and we want to convert that to a ist of tokenized text we can do:
def get_tokens(cord_path):
cord_uid, path = cord_path
if isinstance(path, Path):
tokens = preprocess(load_text(path))
return cord_uid, tokens
return cord_uid, np.nan
cord_tokens = parallel(get_tokens, cord_paths)
Reading and tokenizing each of the over 60,000 JSON files is a very expensive operation, so we want to distribute this load across our CPU cores.
import pandas as pd
metadata = pd.read_csv(PurePath(data_path) / 'metadata.csv')
metadata.describe()
It is a bit limiting, so we will write a function that provides more information than what is available using describe.
from cord.core import describe_dataframe
describe_dataframe(metadata);
ResearchPapers init
Now we are at ResearchPapers.__init__(). The ResearchPapers instance is constructed with a metadata dataframe, which is the dataframe we parsed in load_metadata. A ResearchPapers instance can also be constructed with a subset of the metadata, meaning that we can create an instance containing only COVID-19 related papers, or papers published by Elsevier or any other subset of the metadata that we are interested in. We will show how to use this functionality later.
We can construct the BM25 index from the papers referenced by the metadata. Each metadata row has an abstract column, which contains the abstract of the research paper, which we can preprocess into the index tokens needed by the index. We can, alternatively, create the index tokens from the full text content of the paper, which we load from disk. The full text content gives the potential for a more accurate search, with the tradeoff that it takes a much longer time to build the index. To enable this tradeoff, the load function accepts a parameter index, which deteremines which index strategy to use. ResearchPapers.load(index="text") loaded and indexed from the JSON file contents, while ResearchPapers.load(index"abstract") used the metadata abstract. On a Kaggle instance it took approximately 100 seconds to index from the abstracts versus over 2,000 seconds to index from the JSON texts, though it was about three times faster for each operation on my local laptop.
To save on the time to load from the JSON texts the CORD library, JSON index tokens were processed offline and saved to parquet files. These parquet files were then loaded whenever the option index="text" was selected.
def __init__(self, metadata, data_dir='data', index='abstract', view='html'):
self.data_path = Path(data_dir)
self.num_results = 10
self.view = view
self.metadata = metadata
if 'index_tokens' not in metadata:
print('\nIndexing research papers')
if any([index == t for t in ['text', 'texts', 'content', 'contents']]):
tick = time.time()
_set_index_from_text(self.metadata, data_dir)
print("Finished indexing in", int(time.time() - tick), 'seconds')
else:
print('Creating the BM25 index from the abstracts of the papers')
print('Use index="text" if you want to index the texts of the paper instead')
tick = time.time()
self.metadata['index_tokens'] = metadata.abstract.apply(preprocess)
tock = time.time()
print('Finished Indexing in', round(tock - tick, 0), 'seconds')
# Create BM25 search index
self.bm25 = _get_bm25Okapi(self.metadata.index_tokens)
Python Objects wrapping dataframes
A pattern that is repeated throughout the project is to have a dataframe as a member of a python object. The python object controls access to the dataframe and provides useful functionality, while the dataframe acts as a local database. There are many use cases that fit this pattern where you load read-only data into a dataframe and wrap python code around it. In our case the ResearchPapers class acts like a miniature application that accesses a local in-memory database. We also do this with the SearchResults object, which wraps a dataframe of the search results.
The most important line above self.metadata = metadata sets the dataframe as an instance member. The primary use is to load all the research papers from the metadata.csv, in which case we will have all 50,000+ research papers. Note that we can pass a metadata dataframe of any number of records, meaning that we can make subsets of ResearchPapers. This enables the ability to select only a subset of research papers, e.g. source=="Elsevier", then make a copy of the ResearchPapers with the smaller number of records.
def query(self, query):
data = self.metadata.query(query)
return self._make_copy(data)
def _make_copy(self, new_data):
return ResearchPapers(metadata=new_data.copy(),
data_dir=self.data_path,
view=self.view)
We will see how this pattern is used in more detail later in the notebook.
from cord import ResearchPapers
papers = ResearchPapers.load()
papers = ResearchPapers.load(index='text')
papers
The Search Index
The search index is built on a BM25 Okapi libray called _rankbm25.
An implementation of BM25 is also available in gensim, but I found _rankbm25 was simple to implement.
What is BM25 Okapi?
BM25 stands for Best Match the 25th version and is a text search algorithm first developed in 1994. Okapi stands for the Okapi information retrieval system, implemented at London's City University in the 1980s and 1990s on which BM25 was used.
It is one of the best search algorithms available, and Lucene and its derivatives, Solr and ElasticSearch switched to a BM25 variant around 2015.
Creating the BM25 Index
The heart of this notebook, the one forked over 400 times on Kaggle, is this line of code.
BM25Okapi(index_tokens.tolist())
This creates a new BM25Okapi index on the index_tokens. The index accepts a list of tokens, with each item in the list representing a single tokenized research paper. This is as simple as you can get for a bit of technology that is also used in ElasticSearch software.
The actual implementation in the library is just slightly more complicated, accounting for the edge case of creating a ResearchPapers instance with no papers inside. (This can happen, as we see below, if our API is so flexible that it allows a user to do this).
from rank_bm25 import BM25Okapi
def _get_bm25Okapi(index_tokens):
has_tokens = index_tokens.apply(len).sum() > 0
if not has_tokens:
index_tokens.loc[0] = ['no', 'tokens']
return BM25Okapi(index_tokens.tolist())
Searching Papers
The search function is defined below. We preprocess the search string and then get the doc_scores (the search relevance score) from the BM25 index for all documents in the index. Then we get the dataframe locations of the most relevant papers, do some additional filtering, then return the top n results.
def search(self, search_string,
num_results=10,
covid_related=False,
start_date=None,
end_date=None,
view='html'):
n_results = num_results or self.num_results
# Preprocess the search string
search_terms = preprocess(search_string)
# Get the doc scores from the BM25 index
doc_scores = self.bm25.get_scores(search_terms)
# Get the index from the doc scores sorted by most relevant
ind = np.argsort(doc_scores)[::-1]
# Sort the metadata using the sorted index above
results = self.metadata.iloc[ind].copy()
# Round the doc scores, in case we use the score in the display
results['Score'] = doc_scores[ind].round(1)
# Filter covid related
if covid_related:
results = results[results.covid_related]
# Filter by dates - start date
if start_date:
results = results[results.published >= start_date]
# end data
if end_date:
results = results[results.published < end_date]
# Show only up to n_results
results = results.head(num_results)
# Create the final results
results = results.drop_duplicates(subset=['title'])
# Return Search Results
return SearchResults(results, self.data_path, view=view)
Here is the code run outside of the function, with print statements
from cord.text import preprocess
import numpy as np
search_query = 'Mother to child transmission'
search_tokens = preprocess(search_query)
print('search_tokens', search_tokens)
doc_scores = papers.bm25.get_scores(search_tokens)
print('doc_scores', doc_scores)
ind = np.argsort(doc_scores)[::-1]
print('ind', ind)
The Similarity Index
Included with the CORD-19 Research Dataset is a CSV file containing the document embeddings vectors for the papers in the dataset. Each vector is a 768-dimension vector representing what a neural network has learned about that research paper, and, effectively, it is a signature or a fingerprint of that research paper. This vector can be used to build our similarity index and allow us to find similar papers for any given research paper.
The embeddings CSV file is large—over 700MB in the real dataset. For this notebook we have created a smaller CSV file with only the embeddings for the research papers included with the notebook. If you load and look at the embeddings, you will see 769 columns, one for each of the 768-dimension vector, plus the cord_uid. (The cord_uid is the unique identifier for a research paper in the dataset.)
embeddings_path = data_path / 'cord_19_embeddings.csv'
embeddings = pd.read_csv(embeddings_path)
display(embeddings.shape)
## Look at the first 2 rows and 5 columns
embeddings.iloc[:2, :5]
Let us load the data again, but this time add the column names
VECTOR_COLS = [str(i) for i in range(768)]
COLUMNS = ['cord_uid'] + VECTOR_COLS
embeddings = pd.read_csv(embeddings_path, names=COLUMNS).set_index('cord_uid')
print('Loaded embeddings of shape', embeddings.shape)
## Look at the first 2 rows and 5 columns
embeddings.iloc[:2, :5]
Building the Annoy Index
Annoy is a library for building an index of vectors so that the nearest neighbors to that vector can be easily found. This library is used at Spotify to find music recommendations, so it is one of the best technologies we can possibly use to find similar papers.
To build an annoy index, you first create it with the vector size that you want to store in the index, then add each vector to the index, then build.
from annoy import AnnoyIndex
import random
VECTOR_SIZE = 40
annoy_index = AnnoyIndex(VECTOR_SIZE, 'angular') # Length of item vector that will be indexed
for i, vector in enumerate(vectors):
annoy_index.add_item(i, vector)
annoy_index.build(10) # 10 trees
annoy_index.save('test.ann')
Reduce Vector Dimensions
For the index we wanted to create, we thought 768 dimensions was too much, especially since it meant that the resulting index would be very large on disk, and also too large to fit into a Git repository. Therefore we used PCA to reduce the dimensions to 192, and those 192 dimensional vectors are stored in the Annoy Index.
from sklearn.decomposition import PCA
def downsample(docvectors, dimensions=2):
print(f'Downsampling to {dimensions}D embeddings')
pca = PCA(n_components=dimensions, svd_solver='full')
docvectors_downsampled = pca.fit_transform(docvectors)
return np.squeeze(docvectors_downsampled), pca
Getting Similar Vectors
Once we have the annoy index, we use it to find similar papers using the similar papers function. Annoy has two functions for finding similar papers: get_nns_by_item and get_nns_by_vector. In our function we use get_nns_by_item.
def similar_papers(paper_id, num_items=config.num_similar_items):
from .vectors import SPECTOR_SIMILARITY_INDEX
index = paper_id if isinstance(paper_id, int) else get_index(paper_id)
if not index:
return []
similar_indexes = SPECTOR_SIMILARITY_INDEX.get_nns_by_item(index, num_items, search_k=config.search_k)
similar_cord_uids = document_vectors.iloc[similar_indexes].index.values.tolist()
return [id for id in similar_cord_uids if not id == paper_id]
In the search function we returned a SearchResults object that contained a dataframe with the search results. This is a common pattern used in the notebook—often a dataframe is wrapped inside of an object.
The reason for this pattern is to control how the dataframe is displayed in the notebook. For the search result, we want to present a different formatting from what is available for dataframes, and we may also want to control which columns are displayed, or other aspects of the search results.
Outputting HTML
Objects can control how they are displayed as HTML in a Jupyter notebook by implementing the _repr_html_() function. Some of the objects that we create in this project are meant to wrap a dataframe and control how they are displayed. SearchResults is one such object—it wraps the results dataframe and controls how it looks using _repr_html_(). SearchResults can actually be viewed as a dataframe
papers.search('Mother to child', covid_related=True, num_results=4, view='df')
or an HTML table:
papers.search('Mother to child', covid_related=True, num_results=2)
Using the Dataframe style function
Another great way to customize the look of a dataframe inside a notebook is to use the dataframe.style function. This pandas functionality is pretty powerful and can allow you to completely change how a dataframe is displayed in a notebook.
In the CORD library we wanted to style the results differently from regular dataframes, to improve on the look and feel, and to have the library stand out from the competition. The code below shows how the display function used the style function to change the styling of the results.
def display(self, *paper_ids):
# Look up the papers using the paperids and create a dataframe
_recs = []
for id in paper_ids:
paper = self[id]
_recs.append({'published': paper.metadata.published,
'title': paper.title,
'summary': paper.summary,
'when': paper.metadata.when,
'cord_uid': paper.cord_uid})
df = pd.DataFrame(_recs).sort_values(['published'], ascending=False).drop(columns=['published'])
# Apply a style to a column
def highlight_cols(s):
return 'font-size: 1.1em; color: #008B8B; font-weight: bold'
# Apply the style above to the title column of the dataframe and hide the index
return df.style.applymap(highlight_cols, subset=pd.IndexSlice[:, ['title']]).hide_index()
This is how the results are dispayed in the notebook:
papers.display('v3lbrzh8')
For more information on styling dataframes see Pandas Styling.
Creating Search Widgets
To create interactivity within the notebook, and to allow the user to perform interactive searches, we use ipywidgets.
Search Date Slider
This widget displays a slider that allows a user to select relevant dates for the Research paper search:
def SearchDatesSlider():
options = [(' 1951 ', '1951-01-01'), (' SARS 2003 ', '2002-11-01'),
(' H1N1 2009 ', '2009-04-01'), (' COVID 19 ', '2019-11-30'),
(' 2020 ', '2020-12-31')]
return widgets.SelectionRangeSlider(
options=options,
description='Dates',
disabled=False,
value=('2002-11-01', '2020-12-31'),
layout={'width': '480px'}
)
SearchBar
For the search bar we add a Text, Button, Checkbox and use HBox and VBox for layout.
def searchbar(self, initial_search_terms='', num_results=10, view=None):
text_input = widgets.Text(layout=widgets.Layout(width='400px'), value=initial_search_terms)
search_button = widgets.Button(description='Search', button_style='primary',
layout=widgets.Layout(width='100px'))
search_box = widgets.HBox(children=[text_input, search_button])
# A COVID-related checkbox
covid_related_CheckBox = widgets.Checkbox(description='Covid-19 related', value=False, disable=False)
checkboxes = widgets.HBox(children=[covid_related_CheckBox])
# A date slider to limit research papers to a date range
search_dates_slider = SearchDatesSlider()
search_widget = widgets.VBox([search_box, search_dates_slider, checkboxes])
output = widgets.Output()
def do_search():
search_terms = text_input.value.strip()
if search_terms and len(search_terms) >= 4:
start_date, end_date = search_dates_slider.value
self._search_papers(output=output, SearchTerms=search_terms, num_results=num_results, view=view,
start_date=start_date, end_date=end_date,
covid_related=covid_related_CheckBox.value)
def button_search_handler(btn):
with output:
clear_output()
do_search()
def text_search_handler(change):
if len(change['new'].split(' ')) != len(change['old'].split(' ')):
do_search()
def date_handler(change):
do_search()
def checkbox_handler(change):
do_search()
search_button.on_click(button_search_handler)
text_input.observe(text_search_handler, names='value')
search_dates_slider.observe(date_handler, names='value')
covid_related_CheckBox.observe(checkbox_handler, names='value')
display(search_widget)
display(output)
# Show the initial terms
if initial_search_terms:
do_search()
Now we can see the search bar in action. It updates as you change the selections in the form
papers.searchbar('Cruise ship', num_results=4)
Subsetting Research Papers
There are many ways to select subsets of research papers including:
- Papers since SARS
research_papers.since_sars() - Papers since SARS-COV-2
research_papers.since_sarscov2() - Papers before SARS
research_papers.before_sars() - Papers before SARS-COV-2
research_papers.before_sarscov2() - Papers before a date
research_papers.before('1989-09-12') - Papers after a date
research_papers.after('1989-09-12') - Papers that contains a string
- Papers that match a string (using regex)
papers.since_sarscov2()
papers.contains('bats', column='title')
papers.match('.*McCloskey, B', column='authors')
The ResearchPapers class implements __get_item__() so a single ResearchPaper can be accessed using the numeric index of the paper as in:
papers[0]
It's preferable to access a paper using the cord_uid since it is a stable identifier. papers["5o38ihe0"] referes to the same research paper
papers['5o38ihe0']
Implementing getitem
The implementation of __getitem__() is simple:
def __getitem__(self, item):
if isinstance(item, int):
paper = self.metadata.iloc[item]
else:
paper = self.metadata[self.metadata.cord_uid == item]
The main view of a research paper is the overview, as shown above. This shows a formatted collection of the paper's important fields. There are other views or attributes of research papers, including:
- Overview: A nicely formatted view of the paper's important fields
- Abstract: The paper's abstract
- Summary: A summary of the paper's abstract using the
TextRankalgorithm - Text: The text in the paper
- HTML: The contents of the paper as somewhat nicely formatted HTML
- Text Summary: The text of the paper, summarized using the
TextRankalgorithm
This is implemented in the following function:
paper = research_papers['asf5c7xu']
def view_paper(ViewPaperAs):
if ViewPaperAs == 'Overview':
display(paper)
elif ViewPaperAs == 'Abstract':
display(paper.abstract)
elif ViewPaperAs == 'Summary of Abstract':
display(paper.summary)
elif ViewPaperAs == 'HTML':
display(paper.html)
elif ViewPaperAs == 'Text':
display(paper.text)
elif ViewPaperAs == 'Summary of Text':
display(paper.text_summary)
interact(view_paper,
ViewPaperAs=['Overview', # Show an overview of the paper's important fields and statistics
'Abstract', # Show the paper's abstract
'Summary of Abstract', # Show a summary of the paper's abstract
'HTML', # Show the paper's contents as (slightly) formatted HTML
'Text', # Show the paper's contents
'Summary of Text' # Show a summary of the paper's content
]
);
Conclusion
In this notebook we showed you how to use search and NLP techniques to build a simple search engine and UI over a set of documents. Hopefully, it can help you create your own solution if you are interested in contributing to the same CORD research paper dataset. However, the code and techniques learned here can be applied to other use cases, so feel free to adapt it to your own purposes.
